Spark MLContext Programming Guide
- Overview
- Spark Shell Example
- Start Spark Shell with SystemML
- Create MLContext
- Hello World
- DataFrame Example
- RDD Example
- Matrix Output
- Univariate Statistics on Haberman Data
- Script Information
- Clearing Scripts and MLContext
- Statistics
- Explain
- Script Creation and ScriptFactory
- ScriptExecutor
- MatrixMetadata
- Matrix Data Conversions and Performance
- Jupyter (PySpark) Notebook Example - Poisson Nonnegative Matrix Factorization
- Spark Shell Example - OLD API
- Zeppelin Notebook Example - Linear Regression Algorithm - OLD API
- ** NOTE: This API is old and has been deprecated. **
- Trigger Spark Startup
- Generate Linear Regression Test Data
- Train using Spark ML Linear Regression Algorithm for Comparison
- Spark ML Linear Regression Summary Statistics
- SystemML Linear Regression Algorithm
- Helper Methods
- Convert DataFrame to Binary-Block Format
- Train using SystemML Linear Regression Algorithm
- SystemML Linear Regression Summary Statistics
- Jupyter (PySpark) Notebook Example - Poisson Nonnegative Matrix Factorization - OLD API
- ** NOTE: This API is old and has been deprecated. **
- Set up the notebook and download the data
- Use PySpark to load the data in as a Spark DataFrame
- Create a SystemML MLContext object
- Define a kernel for Poisson nonnegative matrix factorization (PNMF) in DML
- Execute the algorithm
- Retrieve the losses during training and plot them
- Recommended Spark Configuration Settings
Overview
The Spark MLContext API offers a programmatic interface for interacting with SystemML from Spark using languages
such as Scala, Java, and Python. As a result, it offers a convenient way to interact with SystemML from the Spark
Shell and from Notebooks such as Jupyter and Zeppelin.
NOTE: A new MLContext API has been redesigned for future SystemML releases. The old API is available in previous versions of SystemML but is deprecated and will be removed soon, so please migrate to the new API.
Spark Shell Example
Start Spark Shell with SystemML
To use SystemML with Spark Shell, the SystemML jar can be referenced using Spark Shell’s --jars option.
spark-shell --executor-memory 4G --driver-memory 4G --jars SystemML.jarCreate MLContext
All primary classes that a user interacts with are located in the org.apache.sysml.api.mlcontext package.
For convenience, we can additionally add a static import of ScriptFactory to shorten the syntax for creating Script objects.
An MLContext object can be created by passing its constructor a reference to the SparkContext. If successful, you
should see a “Welcome to Apache SystemML!” message.
import org.apache.sysml.api.mlcontext._
import org.apache.sysml.api.mlcontext.ScriptFactory._
val ml = new MLContext(sc)scala> import org.apache.sysml.api.mlcontext._
import org.apache.sysml.api.mlcontext._
scala> import org.apache.sysml.api.mlcontext.ScriptFactory._
import org.apache.sysml.api.mlcontext.ScriptFactory._
scala> val ml = new MLContext(sc)
Welcome to Apache SystemML!
ml: org.apache.sysml.api.mlcontext.MLContext = org.apache.sysml.api.mlcontext.MLContext@12139db0Hello World
The ScriptFactory class allows DML and PYDML scripts to be created from Strings, Files, URLs, and InputStreams.
Here, we’ll use the dml method to create a DML “hello world” script based on a String. Notice that the script
reports that it has no inputs or outputs.
We execute the script using MLContext’s execute method, which displays “hello world” to the console.
The execute method returns an MLResults object, which contains no results since the script has
no outputs.
val helloScript = dml("print('hello world')")
ml.execute(helloScript)scala> val helloScript = dml("print('hello world')")
helloScript: org.apache.sysml.api.mlcontext.Script =
Inputs:
None
Outputs:
None
scala> ml.execute(helloScript)
hello world
res0: org.apache.sysml.api.mlcontext.MLResults =
NoneDataFrame Example
For demonstration purposes, we’ll use Spark to create a DataFrame called df of random doubles from 0 to 1 consisting of 10,000 rows and 1,000 columns.
import org.apache.spark.sql._
import org.apache.spark.sql.types.{StructType,StructField,DoubleType}
import scala.util.Random
val numRows = 10000
val numCols = 1000
val data = sc.parallelize(0 to numRows-1).map { _ => Row.fromSeq(Seq.fill(numCols)(Random.nextDouble)) }
val schema = StructType((0 to numCols-1).map { i => StructField("C" + i, DoubleType, true) } )
val df = sqlContext.createDataFrame(data, schema)scala> import org.apache.spark.sql._
import org.apache.spark.sql._
scala> import org.apache.spark.sql.types.{StructType,StructField,DoubleType}
import org.apache.spark.sql.types.{StructType, StructField, DoubleType}
scala> import scala.util.Random
import scala.util.Random
scala> val numRows = 10000
numRows: Int = 10000
scala> val numCols = 1000
numCols: Int = 1000
scala> val data = sc.parallelize(0 to numRows-1).map { _ => Row.fromSeq(Seq.fill(numCols)(Random.nextDouble)) }
data: org.apache.spark.rdd.RDD[org.apache.spark.sql.Row] = MapPartitionsRDD[1] at map at <console>:42
scala> val schema = StructType((0 to numCols-1).map { i => StructField("C" + i, DoubleType, true) } )
schema: org.apache.spark.sql.types.StructType = StructType(StructField(C0,DoubleType,true), StructField(C1,DoubleType,true), StructField(C2,DoubleType,true), StructField(C3,DoubleType,true), StructField(C4,DoubleType,true), StructField(C5,DoubleType,true), StructField(C6,DoubleType,true), StructField(C7,DoubleType,true), StructField(C8,DoubleType,true), StructField(C9,DoubleType,true), StructField(C10,DoubleType,true), StructField(C11,DoubleType,true), StructField(C12,DoubleType,true), StructField(C13,DoubleType,true), StructField(C14,DoubleType,true), StructField(C15,DoubleType,true), StructField(C16,DoubleType,true), StructField(C17,DoubleType,true), StructField(C18,DoubleType,true), StructField(C19,DoubleType,true), StructField(C20,DoubleType,true), StructField(C21,DoubleType,true), ...
scala> val df = sqlContext.createDataFrame(data, schema)
df: org.apache.spark.sql.DataFrame = [C0: double, C1: double, C2: double, C3: double, C4: double, C5: double, C6: double, C7: double, C8: double, C9: double, C10: double, C11: double, C12: double, C13: double, C14: double, C15: double, C16: double, C17: double, C18: double, C19: double, C20: double, C21: double, C22: double, C23: double, C24: double, C25: double, C26: double, C27: double, C28: double, C29: double, C30: double, C31: double, C32: double, C33: double, C34: double, C35: double, C36: double, C37: double, C38: double, C39: double, C40: double, C41: double, C42: double, C43: double, C44: double, C45: double, C46: double, C47: double, C48: double, C49: double, C50: double, C51: double, C52: double, C53: double, C54: double, C55: double, C56: double, C57: double, C58: double, C5...We’ll create a DML script to find the minimum, maximum, and mean values in a matrix. This
script has one input variable, matrix Xin, and three output variables, minOut, maxOut, and meanOut.
For performance, we’ll specify metadata indicating that the matrix has 10,000 rows and 1,000 columns.
We’ll create a DML script using the ScriptFactory dml method with the minMaxMean script String. The
input variable is specified to be our DataFrame df with MatrixMetadata mm. The output
variables are specified to be minOut, maxOut, and meanOut. Notice that inputs are supplied by the
in method, and outputs are supplied by the out method.
We execute the script and obtain the results as a Tuple by calling getTuple on the results, specifying
the types and names of the output variables.
val minMaxMean =
"""
minOut = min(Xin)
maxOut = max(Xin)
meanOut = mean(Xin)
"""
val mm = new MatrixMetadata(numRows, numCols)
val minMaxMeanScript = dml(minMaxMean).in("Xin", df, mm).out("minOut", "maxOut", "meanOut")
val (min, max, mean) = ml.execute(minMaxMeanScript).getTuple[Double, Double, Double]("minOut", "maxOut", "meanOut")scala> val minMaxMean =
| """
| minOut = min(Xin)
| maxOut = max(Xin)
| meanOut = mean(Xin)
| """
minMaxMean: String =
"
minOut = min(Xin)
maxOut = max(Xin)
meanOut = mean(Xin)
"
scala> val mm = new MatrixMetadata(numRows, numCols)
mm: org.apache.sysml.api.mlcontext.MatrixMetadata = rows: 10000, columns: 1000, non-zeros: None, rows per block: None, columns per block: None
scala> val minMaxMeanScript = dml(minMaxMean).in("Xin", df, mm).out("minOut", "maxOut", "meanOut")
minMaxMeanScript: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (DataFrame) Xin: [C0: double, C1: double, C2: double, C3: double, C4: double, C5: double, C6: double, C7: double, ...
Outputs:
[1] minOut
[2] maxOut
[3] meanOut
scala> val (min, max, mean) = ml.execute(minMaxMeanScript).getTuple[Double, Double, Double]("minOut", "maxOut", "meanOut")
min: Double = 2.6257349849956313E-8
max: Double = 0.9999999686609718
mean: Double = 0.49996223966662934Many different types of input and output variables are automatically allowed. These types include
Boolean, Long, Double, String, Array[Array[Double]], RDD<String> and JavaRDD<String>
in CSV (dense) and IJV (sparse) formats, DataFrame, BinaryBlockMatrix, Matrix, and
Frame. RDDs and JavaRDDs are assumed to be CSV format unless MatrixMetadata is supplied indicating
IJV format.
RDD Example
Let’s take a look at an example of input matrices as RDDs in CSV format. We’ll create two 2x2 matrices and input these into a DML script. This script will sum each matrix and create a message based on which sum is greater. We will output the sums and the message.
For fun, we’ll write the script String to a file and then use ScriptFactory’s dmlFromFile method
to create the script object based on the file. We’ll also specify the inputs using a Map, although
we could have also chained together two in methods to specify the same inputs.
val rdd1 = sc.parallelize(Array("1.0,2.0", "3.0,4.0"))
val rdd2 = sc.parallelize(Array("5.0,6.0", "7.0,8.0"))
val sums = """
s1 = sum(m1);
s2 = sum(m2);
if (s1 > s2) {
message = "s1 is greater"
} else if (s2 > s1) {
message = "s2 is greater"
} else {
message = "s1 and s2 are equal"
}
"""
scala.tools.nsc.io.File("sums.dml").writeAll(sums)
val sumScript = dmlFromFile("sums.dml").in(Map("m1"-> rdd1, "m2"-> rdd2)).out("s1", "s2", "message")
val sumResults = ml.execute(sumScript)
val s1 = sumResults.getDouble("s1")
val s2 = sumResults.getDouble("s2")
val message = sumResults.getString("message")scala> val rdd1 = sc.parallelize(Array("1.0,2.0", "3.0,4.0"))
rdd1: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[42] at parallelize at <console>:38
scala> val rdd2 = sc.parallelize(Array("5.0,6.0", "7.0,8.0"))
rdd2: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[43] at parallelize at <console>:38
scala> val sums = """
| s1 = sum(m1);
| s2 = sum(m2);
| if (s1 > s2) {
| message = "s1 is greater"
| } else if (s2 > s1) {
| message = "s2 is greater"
| } else {
| message = "s1 and s2 are equal"
| }
| """
sums: String =
"
s1 = sum(m1);
s2 = sum(m2);
if (s1 > s2) {
message = "s1 is greater"
} else if (s2 > s1) {
message = "s2 is greater"
} else {
message = "s1 and s2 are equal"
}
"
scala> scala.tools.nsc.io.File("sums.dml").writeAll(sums)
scala> val sumScript = dmlFromFile("sums.dml").in(Map("m1"-> rdd1, "m2"-> rdd2)).out("s1", "s2", "message")
sumScript: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (RDD) m1: ParallelCollectionRDD[42] at parallelize at <console>:38
[2] (RDD) m2: ParallelCollectionRDD[43] at parallelize at <console>:38
Outputs:
[1] s1
[2] s2
[3] message
scala> val sumResults = ml.execute(sumScript)
sumResults: org.apache.sysml.api.mlcontext.MLResults =
[1] (Double) s1: 10.0
[2] (Double) s2: 26.0
[3] (String) message: s2 is greater
scala> val s1 = sumResults.getDouble("s1")
s1: Double = 10.0
scala> val s2 = sumResults.getDouble("s2")
s2: Double = 26.0
scala> val message = sumResults.getString("message")
message: String = s2 is greaterIf you have metadata that you would like to supply along with the input matrices, this can be accomplished using a Scala Seq, List, or Array.
val rdd1Metadata = new MatrixMetadata(2, 2)
val rdd2Metadata = new MatrixMetadata(2, 2)
val sumScript = dmlFromFile("sums.dml").in(Seq(("m1", rdd1, rdd1Metadata), ("m2", rdd2, rdd2Metadata))).out("s1", "s2", "message")
val (firstSum, secondSum, sumMessage) = ml.execute(sumScript).getTuple[Double, Double, String]("s1", "s2", "message")scala> val rdd1Metadata = new MatrixMetadata(2, 2)
rdd1Metadata: org.apache.sysml.api.mlcontext.MatrixMetadata = rows: 2, columns: 2, non-zeros: None, rows per block: None, columns per block: None
scala> val rdd2Metadata = new MatrixMetadata(2, 2)
rdd2Metadata: org.apache.sysml.api.mlcontext.MatrixMetadata = rows: 2, columns: 2, non-zeros: None, rows per block: None, columns per block: None
scala> val sumScript = dmlFromFile("sums.dml").in(Seq(("m1", rdd1, rdd1Metadata), ("m2", rdd2, rdd2Metadata))).out("s1", "s2", "message")
sumScript: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (RDD) m1: ParallelCollectionRDD[42] at parallelize at <console>:38
[2] (RDD) m2: ParallelCollectionRDD[43] at parallelize at <console>:38
Outputs:
[1] s1
[2] s2
[3] message
scala> val (firstSum, secondSum, sumMessage) = ml.execute(sumScript).getTuple[Double, Double, String]("s1", "s2", "message")
firstSum: Double = 10.0
secondSum: Double = 26.0
sumMessage: String = s2 is greaterThe same inputs with metadata can be supplied by chaining in methods, as in the example below, which shows that out methods can also be
chained.
val sumScript = dmlFromFile("sums.dml").in("m1", rdd1, rdd1Metadata).in("m2", rdd2, rdd2Metadata).out("s1").out("s2").out("message")
val (firstSum, secondSum, sumMessage) = ml.execute(sumScript).getTuple[Double, Double, String]("s1", "s2", "message")scala> val sumScript = dmlFromFile("sums.dml").in("m1", rdd1, rdd1Metadata).in("m2", rdd2, rdd2Metadata).out("s1").out("s2").out("message")
sumScript: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (RDD) m1: ParallelCollectionRDD[42] at parallelize at <console>:38
[2] (RDD) m2: ParallelCollectionRDD[43] at parallelize at <console>:38
Outputs:
[1] s1
[2] s2
[3] message
scala> val (firstSum, secondSum, sumMessage) = ml.execute(sumScript).getTuple[Double, Double, String]("s1", "s2", "message")
firstSum: Double = 10.0
secondSum: Double = 26.0
sumMessage: String = s2 is greaterMatrix Output
Let’s look at an example of reading a matrix out of SystemML. We’ll create a DML script
in which we create a 2x2 matrix m. We’ll set the variable n to be the sum of the cells in the matrix.
We create a script object using String s, and we set m and n as the outputs. We execute the script, and in
the results we see we have Matrix m and Double n. The n output variable has a value of 110.0.
We get Matrix m and Double n as a Tuple of values x and y. We then convert Matrix m to an
RDD of IJV values, an RDD of CSV values, a DataFrame, and a two-dimensional Double Array, and we display
the values in each of these data structures.
val s =
"""
m = matrix("11 22 33 44", rows=2, cols=2)
n = sum(m)
"""
val scr = dml(s).out("m", "n");
val res = ml.execute(scr)
val (x, y) = res.getTuple[Matrix, Double]("m", "n")
x.toRDDStringIJV.collect.foreach(println)
x.toRDDStringCSV.collect.foreach(println)
x.toDF.collect.foreach(println)
x.to2DDoubleArrayscala> val s =
| """
| m = matrix("11 22 33 44", rows=2, cols=2)
| n = sum(m)
| """
s: String =
"
m = matrix("11 22 33 44", rows=2, cols=2)
n = sum(m)
"
scala> val scr = dml(s).out("m", "n");
scr: org.apache.sysml.api.mlcontext.Script =
Inputs:
None
Outputs:
[1] m
[2] n
scala> val res = ml.execute(scr)
res: org.apache.sysml.api.mlcontext.MLResults =
[1] (Matrix) m: Matrix: scratch_space//_p12059_9.31.117.12//_t0/temp26_14, [2 x 2, nnz=4, blocks (1000 x 1000)], binaryblock, dirty
[2] (Double) n: 110.0
scala> val (x, y) = res.getTuple[Matrix, Double]("m", "n")
x: org.apache.sysml.api.mlcontext.Matrix = Matrix: scratch_space//_p12059_9.31.117.12//_t0/temp26_14, [2 x 2, nnz=4, blocks (1000 x 1000)], binaryblock, dirty
y: Double = 110.0
scala> x.toRDDStringIJV.collect.foreach(println)
1 1 11.0
1 2 22.0
2 1 33.0
2 2 44.0
scala> x.toRDDStringCSV.collect.foreach(println)
11.0,22.0
33.0,44.0
scala> x.toDF.collect.foreach(println)
[0.0,11.0,22.0]
[1.0,33.0,44.0]
scala> x.to2DDoubleArray
res10: Array[Array[Double]] = Array(Array(11.0, 22.0), Array(33.0, 44.0))Univariate Statistics on Haberman Data
Our next example will involve Haberman’s Survival Data Set in CSV format from the Center for Machine Learning and Intelligent Systems. We will run the SystemML Univariate Statistics (“Univar-Stats.dml”) script on this data.
We’ll pull the data from a URL and convert it to an RDD, habermanRDD. Next, we’ll create metadata, habermanMetadata,
stating that the matrix consists of 306 rows and 4 columns.
As we can see from the comments in the script
here, the
script requires a ‘TYPES’ input matrix that lists the types of the features (1 for scale, 2 for nominal, 3 for
ordinal), so we create a typesRDD matrix consisting of 1 row and 4 columns, with corresponding metadata, typesMetadata.
Next, we create the DML script object called uni using ScriptFactory’s dmlFromUrl method, specifying the GitHub URL where the
DML script is located. We bind the habermanRDD matrix to the A variable in Univar-Stats.dml, and we bind
the typesRDD matrix to the K variable. In addition, we supply a $CONSOLE_OUTPUT parameter with a Boolean value
of true, which indicates that we’d like to output labeled results to the console. We’ll explain why we bind to the A and K
variables in the Input Variables vs Input Parameters
section below.
val habermanUrl = "http://archive.ics.uci.edu/ml/machine-learning-databases/haberman/haberman.data"
val habermanList = scala.io.Source.fromURL(habermanUrl).mkString.split("\n")
val habermanRDD = sc.parallelize(habermanList)
val habermanMetadata = new MatrixMetadata(306, 4)
val typesRDD = sc.parallelize(Array("1.0,1.0,1.0,2.0"))
val typesMetadata = new MatrixMetadata(1, 4)
val scriptUrl = "https://raw.githubusercontent.com/apache/incubator-systemml/master/scripts/algorithms/Univar-Stats.dml"
val uni = dmlFromUrl(scriptUrl).in("A", habermanRDD, habermanMetadata).in("K", typesRDD, typesMetadata).in("$CONSOLE_OUTPUT", true)
ml.execute(uni)scala> val habermanUrl = "http://archive.ics.uci.edu/ml/machine-learning-databases/haberman/haberman.data"
habermanUrl: String = http://archive.ics.uci.edu/ml/machine-learning-databases/haberman/haberman.data
scala> val habermanList = scala.io.Source.fromURL(habermanUrl).mkString.split("\n")
habermanList: Array[String] = Array(30,64,1,1, 30,62,3,1, 30,65,0,1, 31,59,2,1, 31,65,4,1, 33,58,10,1, 33,60,0,1, 34,59,0,2, 34,66,9,2, 34,58,30,1, 34,60,1,1, 34,61,10,1, 34,67,7,1, 34,60,0,1, 35,64,13,1, 35,63,0,1, 36,60,1,1, 36,69,0,1, 37,60,0,1, 37,63,0,1, 37,58,0,1, 37,59,6,1, 37,60,15,1, 37,63,0,1, 38,69,21,2, 38,59,2,1, 38,60,0,1, 38,60,0,1, 38,62,3,1, 38,64,1,1, 38,66,0,1, 38,66,11,1, 38,60,1,1, 38,67,5,1, 39,66,0,2, 39,63,0,1, 39,67,0,1, 39,58,0,1, 39,59,2,1, 39,63,4,1, 40,58,2,1, 40,58,0,1, 40,65,0,1, 41,60,23,2, 41,64,0,2, 41,67,0,2, 41,58,0,1, 41,59,8,1, 41,59,0,1, 41,64,0,1, 41,69,8,1, 41,65,0,1, 41,65,0,1, 42,69,1,2, 42,59,0,2, 42,58,0,1, 42,60,1,1, 42,59,2,1, 42,61,4,1, 42,62,20,1, 42,65,0,1, 42,63,1,1, 43,58,52,2, 43,59,2,2, 43,64,0,2, 43,64,0,2, 43,63,14,1, 43,64,2,1, 43...
scala> val habermanRDD = sc.parallelize(habermanList)
habermanRDD: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[159] at parallelize at <console>:43
scala> val habermanMetadata = new MatrixMetadata(306, 4)
habermanMetadata: org.apache.sysml.api.mlcontext.MatrixMetadata = rows: 306, columns: 4, non-zeros: None, rows per block: None, columns per block: None
scala> val typesRDD = sc.parallelize(Array("1.0,1.0,1.0,2.0"))
typesRDD: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[160] at parallelize at <console>:39
scala> val typesMetadata = new MatrixMetadata(1, 4)
typesMetadata: org.apache.sysml.api.mlcontext.MatrixMetadata = rows: 1, columns: 4, non-zeros: None, rows per block: None, columns per block: None
scala> val scriptUrl = "https://raw.githubusercontent.com/apache/incubator-systemml/master/scripts/algorithms/Univar-Stats.dml"
scriptUrl: String = https://raw.githubusercontent.com/apache/incubator-systemml/master/scripts/algorithms/Univar-Stats.dml
scala> val uni = dmlFromUrl(scriptUrl).in("A", habermanRDD, habermanMetadata).in("K", typesRDD, typesMetadata).in("$CONSOLE_OUTPUT", true)
uni: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (RDD) A: ParallelCollectionRDD[159] at parallelize at <console>:43
[2] (RDD) K: ParallelCollectionRDD[160] at parallelize at <console>:39
[3] (Boolean) $CONSOLE_OUTPUT: true
Outputs:
None
scala> ml.execute(uni)
...
-------------------------------------------------
Feature [1]: Scale
(01) Minimum | 30.0
(02) Maximum | 83.0
(03) Range | 53.0
(04) Mean | 52.45751633986928
(05) Variance | 116.71458266366658
(06) Std deviation | 10.803452349303281
(07) Std err of mean | 0.6175922641866753
(08) Coeff of variation | 0.20594669940735139
(09) Skewness | 0.1450718616532357
(10) Kurtosis | -0.6150152487211726
(11) Std err of skewness | 0.13934809593495995
(12) Std err of kurtosis | 0.277810485320835
(13) Median | 52.0
(14) Interquartile mean | 52.16013071895425
-------------------------------------------------
Feature [2]: Scale
(01) Minimum | 58.0
(02) Maximum | 69.0
(03) Range | 11.0
(04) Mean | 62.85294117647059
(05) Variance | 10.558630665380907
(06) Std deviation | 3.2494046632238507
(07) Std err of mean | 0.18575610076612029
(08) Coeff of variation | 0.051698529971741194
(09) Skewness | 0.07798443581479181
(10) Kurtosis | -1.1324380182967442
(11) Std err of skewness | 0.13934809593495995
(12) Std err of kurtosis | 0.277810485320835
(13) Median | 63.0
(14) Interquartile mean | 62.80392156862745
-------------------------------------------------
Feature [3]: Scale
(01) Minimum | 0.0
(02) Maximum | 52.0
(03) Range | 52.0
(04) Mean | 4.026143790849673
(05) Variance | 51.691117539912135
(06) Std deviation | 7.189653506248555
(07) Std err of mean | 0.41100513466216837
(08) Coeff of variation | 1.7857418611299172
(09) Skewness | 2.954633471088322
(10) Kurtosis | 11.425776549251449
(11) Std err of skewness | 0.13934809593495995
(12) Std err of kurtosis | 0.277810485320835
(13) Median | 1.0
(14) Interquartile mean | 1.2483660130718954
-------------------------------------------------
Feature [4]: Categorical (Nominal)
(15) Num of categories | 2
(16) Mode | 1
(17) Num of modes | 1
res23: org.apache.sysml.api.mlcontext.MLResults =
NoneAlternatively, we could supply a java.net.URL to the Script in method. Note that if the URL matrix data is in IJV
format, metadata needs to be supplied for the matrix.
val habermanUrl = "http://archive.ics.uci.edu/ml/machine-learning-databases/haberman/haberman.data"
val typesRDD = sc.parallelize(Array("1.0,1.0,1.0,2.0"))
val scriptUrl = "https://raw.githubusercontent.com/apache/incubator-systemml/master/scripts/algorithms/Univar-Stats.dml"
val uni = dmlFromUrl(scriptUrl).in("A", new java.net.URL(habermanUrl)).in("K", typesRDD).in("$CONSOLE_OUTPUT", true)
ml.execute(uni)scala> val habermanUrl = "http://archive.ics.uci.edu/ml/machine-learning-databases/haberman/haberman.data"
habermanUrl: String = http://archive.ics.uci.edu/ml/machine-learning-databases/haberman/haberman.data
scala> val typesRDD = sc.parallelize(Array("1.0,1.0,1.0,2.0"))
typesRDD: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[50] at parallelize at <console>:33
scala> val scriptUrl = "https://raw.githubusercontent.com/apache/incubator-systemml/master/scripts/algorithms/Univar-Stats.dml"
scriptUrl: String = https://raw.githubusercontent.com/apache/incubator-systemml/master/scripts/algorithms/Univar-Stats.dml
scala> val uni = dmlFromUrl(scriptUrl).in("A", new java.net.URL(habermanUrl)).in("K", typesRDD).in("$CONSOLE_OUTPUT", true)
uni: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (URL) A: http://archive.ics.uci.edu/ml/machine-learning-databases/haberman/haberman.data
[2] (RDD) K: ParallelCollectionRDD[50] at parallelize at <console>:33
[3] (Boolean) $CONSOLE_OUTPUT: true
Outputs:
None
scala> ml.execute(uni)
...
-------------------------------------------------
(01) Minimum | 30.0
(02) Maximum | 83.0
(03) Range | 53.0
(04) Mean | 52.45751633986928
(05) Variance | 116.71458266366658
(06) Std deviation | 10.803452349303281
(07) Std err of mean | 0.6175922641866753
(08) Coeff of variation | 0.20594669940735139
(09) Skewness | 0.1450718616532357
(10) Kurtosis | -0.6150152487211726
(11) Std err of skewness | 0.13934809593495995
(12) Std err of kurtosis | 0.277810485320835
(13) Median | 52.0
(14) Interquartile mean | 52.16013071895425
Feature [1]: Scale
-------------------------------------------------
(01) Minimum | 58.0
(02) Maximum | 69.0
(03) Range | 11.0
(04) Mean | 62.85294117647059
(05) Variance | 10.558630665380907
(06) Std deviation | 3.2494046632238507
(07) Std err of mean | 0.18575610076612029
(08) Coeff of variation | 0.051698529971741194
(09) Skewness | 0.07798443581479181
(10) Kurtosis | -1.1324380182967442
(11) Std err of skewness | 0.13934809593495995
(12) Std err of kurtosis | 0.277810485320835
(13) Median | 63.0
(14) Interquartile mean | 62.80392156862745
Feature [2]: Scale
-------------------------------------------------
(01) Minimum | 0.0
(02) Maximum | 52.0
(03) Range | 52.0
(04) Mean | 4.026143790849673
(05) Variance | 51.691117539912135
(06) Std deviation | 7.189653506248555
(07) Std err of mean | 0.41100513466216837
(08) Coeff of variation | 1.7857418611299172
(09) Skewness | 2.954633471088322
(10) Kurtosis | 11.425776549251449
(11) Std err of skewness | 0.13934809593495995
(12) Std err of kurtosis | 0.277810485320835
(13) Median | 1.0
(14) Interquartile mean | 1.2483660130718954
Feature [3]: Scale
-------------------------------------------------
Feature [4]: Categorical (Nominal)
(15) Num of categories | 2
(16) Mode | 1
(17) Num of modes | 1
res5: org.apache.sysml.api.mlcontext.MLResults =
NoneInput Variables vs Input Parameters
If we examine the
Univar-Stats.dml
file, we see in the comments that it can take 4 input
parameters, $X, $TYPES, $CONSOLE_OUTPUT, and $STATS. Input parameters are typically useful when
executing SystemML in Standalone mode, Spark batch mode, or Hadoop batch mode. For example, $X specifies
the location in the file system where the input data matrix is located, $TYPES specifies the location in the file system
where the input types matrix is located, $CONSOLE_OUTPUT specifies whether or not labeled statistics should be
output to the console, and $STATS specifies the location in the file system where the output matrix should be written.
...
# INPUT PARAMETERS:
# -------------------------------------------------------------------------------------------------
# NAME TYPE DEFAULT MEANING
# -------------------------------------------------------------------------------------------------
# X String --- Location of INPUT data matrix
# TYPES String --- Location of INPUT matrix that lists the types of the features:
# 1 for scale, 2 for nominal, 3 for ordinal
# CONSOLE_OUTPUT Boolean FALSE If TRUE, print summary statistics to console
# STATS String --- Location of OUTPUT matrix with summary statistics computed for
# all features (17 statistics - 14 scale, 3 categorical)
# -------------------------------------------------------------------------------------------------
# OUTPUT: Matrix of summary statistics
...
consoleOutput = ifdef($CONSOLE_OUTPUT, FALSE);
A = read($X); # data file
K = read($TYPES); # attribute kind file
...
write(baseStats, $STATS);
...Because MLContext is a programmatic interface, it offers more flexibility. You can still use input parameters and files in the file system, such as this example that specifies file paths to the input matrices and the output matrix:
val script = dmlFromFile("scripts/algorithms/Univar-Stats.dml").in("$X", "data/haberman.data").in("$TYPES", "data/types.csv").in("$STATS", "data/univarOut.mtx").in("$CONSOLE_OUTPUT", true)
ml.execute(script)Using the MLContext API, rather than relying solely on input parameters, we can bind to the variables associated
with the read and write statements. In the fragment of Univar-Stats.dml above, notice that the matrix at
path $X is read to variable A, $TYPES is read to variable
K, and baseStats is written to path $STATS. Therefore, we can bind the Haberman input data matrix to the A variable,
the input types matrix to the K variable, and the output matrix to the baseStats variable.
val uni = dmlFromUrl(scriptUrl).in("A", habermanRDD, habermanMetadata).in("K", typesRDD, typesMetadata).out("baseStats")
val baseStats = ml.execute(uni).getMatrix("baseStats")
baseStats.toRDDStringIJV.collect.slice(0,9).foreach(println)scala> val uni = dmlFromUrl(scriptUrl).in("A", habermanRDD, habermanMetadata).in("K", typesRDD, typesMetadata).out("baseStats")
uni: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (RDD) A: ParallelCollectionRDD[159] at parallelize at <console>:43
[2] (RDD) K: ParallelCollectionRDD[160] at parallelize at <console>:39
Outputs:
[1] baseStats
scala> val baseStats = ml.execute(uni).getMatrix("baseStats")
...
baseStats: org.apache.sysml.api.mlcontext.Matrix = Matrix: scratch_space/_p12059_9.31.117.12/parfor/4_resultmerge1, [17 x 4, nnz=44, blocks (1000 x 1000)], binaryblock, dirty
scala> baseStats.toRDDStringIJV.collect.slice(0,9).foreach(println)
1 1 30.0
1 2 58.0
1 3 0.0
1 4 0.0
2 1 83.0
2 2 69.0
2 3 52.0
2 4 0.0
3 1 53.0Script Information
The info method on a Script object can provide useful information about a DML or PyDML script, such as
the inputs, output, symbol table, script string, and the script execution string that is passed to the internals of
SystemML.
val minMaxMean =
"""
minOut = min(Xin)
maxOut = max(Xin)
meanOut = mean(Xin)
"""
val minMaxMeanScript = dml(minMaxMean).in("Xin", df, mm).out("minOut", "maxOut", "meanOut")
val (min, max, mean) = ml.execute(minMaxMeanScript).getTuple[Double, Double, Double]("minOut", "maxOut", "meanOut")
println(minMaxMeanScript.info)scala> val minMaxMean =
| """
| minOut = min(Xin)
| maxOut = max(Xin)
| meanOut = mean(Xin)
| """
minMaxMean: String =
"
minOut = min(Xin)
maxOut = max(Xin)
meanOut = mean(Xin)
"
scala> val minMaxMeanScript = dml(minMaxMean).in("Xin", df, mm).out("minOut", "maxOut", "meanOut")
minMaxMeanScript: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (DataFrame) Xin: [C0: double, C1: double, C2: double, C3: double, C4: double, C5: double, C6: double, C7: double, ...
Outputs:
[1] minOut
[2] maxOut
[3] meanOut
scala> val (min, max, mean) = ml.execute(minMaxMeanScript).getTuple[Double, Double, Double]("minOut", "maxOut", "meanOut")
min: Double = 1.4149740823476975E-7
max: Double = 0.9999999956646207
mean: Double = 0.5000954668004209
scala> println(minMaxMeanScript.info)
Script Type: DML
Inputs:
[1] (DataFrame) Xin: [C0: double, C1: double, C2: double, C3: double, C4: double, C5: double, C6: double, C7: double, ...
Outputs:
[1] (Double) minOut: 1.4149740823476975E-7
[2] (Double) maxOut: 0.9999999956646207
[3] (Double) meanOut: 0.5000954668004209
Input Parameters:
None
Input Variables:
[1] Xin
Output Variables:
[1] minOut
[2] maxOut
[3] meanOut
Symbol Table:
[1] (Double) meanOut: 0.5000954668004209
[2] (Double) maxOut: 0.9999999956646207
[3] (Double) minOut: 1.4149740823476975E-7
[4] (Matrix) Xin: Matrix: scratch_space/temp_1166464711339222, [10000 x 1000, nnz=10000000, blocks (1000 x 1000)], binaryblock, not-dirty
Script String:
minOut = min(Xin)
maxOut = max(Xin)
meanOut = mean(Xin)
Script Execution String:
Xin = read('');
minOut = min(Xin)
maxOut = max(Xin)
meanOut = mean(Xin)
write(minOut, '');
write(maxOut, '');
write(meanOut, '');Clearing Scripts and MLContext
Dealing with large matrices can require a significant amount of memory. To deal help deal with this, you
can call a Script object’s clearAll method to clear the inputs, outputs, symbol table, and script string.
In terms of memory, the symbol table is most important because it holds references to matrices.
In this example, we display the symbol table of the minMaxMeanScript, call clearAll on the script, and
then display the symbol table, which is empty.
println(minMaxMeanScript.displaySymbolTable)
minMaxMeanScript.clearAll
println(minMaxMeanScript.displaySymbolTable)scala> println(minMaxMeanScript.displaySymbolTable)
Symbol Table:
[1] (Double) meanOut: 0.5000954668004209
[2] (Double) maxOut: 0.9999999956646207
[3] (Double) minOut: 1.4149740823476975E-7
[4] (Matrix) Xin: Matrix: scratch_space/temp_1166464711339222, [10000 x 1000, nnz=10000000, blocks (1000 x 1000)], binaryblock, not-dirty
scala> minMaxMeanScript.clearAll
scala> println(minMaxMeanScript.displaySymbolTable)
Symbol Table:
NoneThe MLContext object holds references to the scripts that have been executed. Calling clear on
the MLContext clears all scripts that it has references to and then removes the references to these
scripts.
ml.clearStatistics
Statistics about script executions can be output to the console by calling MLContext’s setStatistics
method with a value of true.
ml.setStatistics(true)
val minMaxMean =
"""
minOut = min(Xin)
maxOut = max(Xin)
meanOut = mean(Xin)
"""
val minMaxMeanScript = dml(minMaxMean).in("Xin", df, mm).out("minOut", "maxOut", "meanOut")
val (min, max, mean) = ml.execute(minMaxMeanScript).getTuple[Double, Double, Double]("minOut", "maxOut", "meanOut")scala> ml.setStatistics(true)
scala> val minMaxMean =
| """
| minOut = min(Xin)
| maxOut = max(Xin)
| meanOut = mean(Xin)
| """
minMaxMean: String =
"
minOut = min(Xin)
maxOut = max(Xin)
meanOut = mean(Xin)
"
scala> val minMaxMeanScript = dml(minMaxMean).in("Xin", df, mm).out("minOut", "maxOut", "meanOut")
minMaxMeanScript: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (DataFrame) Xin: [C0: double, C1: double, C2: double, C3: double, C4: double, C5: double, C6: double, C7: double, ...
Outputs:
[1] minOut
[2] maxOut
[3] meanOut
scala> val (min, max, mean) = ml.execute(minMaxMeanScript).getTuple[Double, Double, Double]("minOut", "maxOut", "meanOut")
SystemML Statistics:
Total elapsed time: 0.000 sec.
Total compilation time: 0.000 sec.
Total execution time: 0.000 sec.
Number of compiled Spark inst: 0.
Number of executed Spark inst: 0.
Cache hits (Mem, WB, FS, HDFS): 2/0/0/1.
Cache writes (WB, FS, HDFS): 1/0/0.
Cache times (ACQr/m, RLS, EXP): 3.137/0.000/0.001/0.000 sec.
HOP DAGs recompiled (PRED, SB): 0/0.
HOP DAGs recompile time: 0.000 sec.
Spark ctx create time (lazy): 0.000 sec.
Spark trans counts (par,bc,col):0/0/2.
Spark trans times (par,bc,col): 0.000/0.000/6.434 secs.
Total JIT compile time: 112.372 sec.
Total JVM GC count: 54.
Total JVM GC time: 9.664 sec.
Heavy hitter instructions (name, time, count):
-- 1) uamin 3.150 sec 1
-- 2) uamean 0.021 sec 1
-- 3) uamax 0.017 sec 1
-- 4) rmvar 0.000 sec 3
-- 5) assignvar 0.000 sec 3
min: Double = 2.4982850344024143E-8
max: Double = 0.9999997007231808
mean: Double = 0.5002109404821844Explain
A DML or PyDML script is converted into a SystemML program during script execution. Information
about this program can be displayed by calling MLContext’s setExplain method with a value
of true.
ml.setExplain(true)
val minMaxMean =
"""
minOut = min(Xin)
maxOut = max(Xin)
meanOut = mean(Xin)
"""
val mm = new MatrixMetadata(numRows, numCols)
val minMaxMeanScript = dml(minMaxMean).in("Xin", df, mm).out("minOut", "maxOut", "meanOut")
val (min, max, mean) = ml.execute(minMaxMeanScript).getTuple[Double, Double, Double]("minOut", "maxOut", "meanOut")scala> ml.setExplain(true)
scala> val minMaxMean =
| """
| minOut = min(Xin)
| maxOut = max(Xin)
| meanOut = mean(Xin)
| """
minMaxMean: String =
"
minOut = min(Xin)
maxOut = max(Xin)
meanOut = mean(Xin)
"
scala> val mm = new MatrixMetadata(numRows, numCols)
mm: org.apache.sysml.api.mlcontext.MatrixMetadata = rows: 10000, columns: 1000, non-zeros: None, rows per block: None, columns per block: None
scala> val minMaxMeanScript = dml(minMaxMean).in("Xin", df, mm).out("minOut", "maxOut", "meanOut")
minMaxMeanScript: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (DataFrame) Xin: [C0: double, C1: double, C2: double, C3: double, C4: double, C5: double, C6: double, C7: double, ...
Outputs:
[1] minOut
[2] maxOut
[3] meanOut
scala> val (min, max, mean) = ml.execute(minMaxMeanScript).getTuple[Double, Double, Double]("minOut", "maxOut", "meanOut")
PROGRAM
--MAIN PROGRAM
----GENERIC (lines 1-8) [recompile=false]
------(12) TRead Xin [10000,1000,1000,1000,10000000] [0,0,76 -> 76MB] [chkpt], CP
------(13) ua(minRC) (12) [0,0,-1,-1,-1] [76,0,0 -> 76MB], CP
------(21) TWrite minOut (13) [0,0,-1,-1,-1] [0,0,0 -> 0MB], CP
------(14) ua(maxRC) (12) [0,0,-1,-1,-1] [76,0,0 -> 76MB], CP
------(27) TWrite maxOut (14) [0,0,-1,-1,-1] [0,0,0 -> 0MB], CP
------(15) ua(meanRC) (12) [0,0,-1,-1,-1] [76,0,0 -> 76MB], CP
------(33) TWrite meanOut (15) [0,0,-1,-1,-1] [0,0,0 -> 0MB], CP
min: Double = 5.16651366133658E-9
max: Double = 0.9999999368927975
mean: Double = 0.5001096515241128Different explain levels can be set. The explain levels are NONE, HOPS, RUNTIME, RECOMPILE_HOPS, and RECOMPILE_RUNTIME.
ml.setExplainLevel(MLContext.ExplainLevel.RUNTIME)
val (min, max, mean) = ml.execute(minMaxMeanScript).getTuple[Double, Double, Double]("minOut", "maxOut", "meanOut")scala> ml.setExplainLevel(MLContext.ExplainLevel.RUNTIME)
scala> val (min, max, mean) = ml.execute(minMaxMeanScript).getTuple[Double, Double, Double]("minOut", "maxOut", "meanOut")
PROGRAM ( size CP/SP = 9/0 )
--MAIN PROGRAM
----GENERIC (lines 1-8) [recompile=false]
------CP uamin Xin.MATRIX.DOUBLE _Var8.SCALAR.DOUBLE 8
------CP uamax Xin.MATRIX.DOUBLE _Var9.SCALAR.DOUBLE 8
------CP uamean Xin.MATRIX.DOUBLE _Var10.SCALAR.DOUBLE 8
------CP assignvar _Var8.SCALAR.DOUBLE.false minOut.SCALAR.DOUBLE
------CP assignvar _Var9.SCALAR.DOUBLE.false maxOut.SCALAR.DOUBLE
------CP assignvar _Var10.SCALAR.DOUBLE.false meanOut.SCALAR.DOUBLE
------CP rmvar _Var8
------CP rmvar _Var9
------CP rmvar _Var10
min: Double = 5.16651366133658E-9
max: Double = 0.9999999368927975
mean: Double = 0.5001096515241128Script Creation and ScriptFactory
Script objects can be created using standard Script constructors. A Script can be of two types: DML (R-based syntax) and PYDML (Python-based syntax). If no ScriptType is specified, the default Script type is DML.
val script = new Script()
println(script.getScriptType)
val script = new Script(ScriptType.PYDML)
println(script.getScriptType)scala> val script = new Script();
...
scala> println(script.getScriptType)
DML
scala> val script = new Script(ScriptType.PYDML);
...
scala> println(script.getScriptType)
PYDMLThe ScriptFactory class offers convenient methods for creating DML and PYDML scripts from a variety of sources. ScriptFactory can create a script object from a String, File, URL, or InputStream.
Script from URL:
Here we create Script object s1 by reading Univar-Stats.dml from a URL.
val uniUrl = "https://raw.githubusercontent.com/apache/incubator-systemml/master/scripts/algorithms/Univar-Stats.dml"
val s1 = ScriptFactory.dmlFromUrl(scriptUrl)Script from String:
We create Script objects s2 and s3 from Strings using ScriptFactory’s dml and dmlFromString methods.
Both methods perform the same action. This example reads an algorithm at a URL to String uniString and then
creates two script objects based on this String.
val uniUrl = "https://raw.githubusercontent.com/apache/incubator-systemml/master/scripts/algorithms/Univar-Stats.dml"
val uniString = scala.io.Source.fromURL(uniUrl).mkString
val s2 = ScriptFactory.dml(uniString)
val s3 = ScriptFactory.dmlFromString(uniString)Script from File:
We create Script object s4 based on a path to a file using ScriptFactory’s dmlFromFile method. This example
reads a URL to a String, writes this String to a file, and then uses the path to the file to create a Script object.
val uniUrl = "https://raw.githubusercontent.com/apache/incubator-systemml/master/scripts/algorithms/Univar-Stats.dml"
val uniString = scala.io.Source.fromURL(uniUrl).mkString
scala.tools.nsc.io.File("uni.dml").writeAll(uniString)
val s4 = ScriptFactory.dmlFromFile("uni.dml")Script from InputStream:
The SystemML jar file contains all the primary algorithm scripts. We can read one of these scripts as an InputStream and use this to create a Script object.
val inputStream = getClass.getResourceAsStream("/scripts/algorithms/Univar-Stats.dml")
val s5 = ScriptFactory.dmlFromInputStream(inputStream)Script from Resource:
As mentioned, the SystemML jar file contains all the primary algorithm script files. For convenience, we can
read these script files or other script files on the classpath using ScriptFactory’s dmlFromResource and pydmlFromResource
methods.
val s6 = ScriptFactory.dmlFromResource("/scripts/algorithms/Univar-Stats.dml");ScriptExecutor
A Script is executed by a ScriptExecutor. If no ScriptExecutor is specified, a default ScriptExecutor will be created to execute a Script. Script execution consists of several steps, as detailed in SystemML’s Optimizer: Plan Generation for Large-Scale Machine Learning Programs. Additional information can be found in the Javadocs for ScriptExecutor.
Advanced users may find it useful to be able to specify their own execution or to override ScriptExecutor methods by subclassing ScriptExecutor.
In this example, we override the parseScript and validateScript methods to display messages to the console
during these execution steps.
class MyScriptExecutor extends org.apache.sysml.api.mlcontext.ScriptExecutor {
override def parseScript{ println("Parsing script"); super.parseScript(); }
override def validateScript{ println("Validating script"); super.validateScript(); }
}
val helloScript = dml("print('hello world')")
ml.execute(helloScript, new MyScriptExecutor)scala> class MyScriptExecutor extends org.apache.sysml.api.mlcontext.ScriptExecutor {
| override def parseScript{ println("Parsing script"); super.parseScript(); }
| override def validateScript{ println("Validating script"); super.validateScript(); }
| }
defined class MyScriptExecutor
scala> val helloScript = dml("print('hello world')")
helloScript: org.apache.sysml.api.mlcontext.Script =
Inputs:
None
Outputs:
None
scala> ml.execute(helloScript, new MyScriptExecutor)
Parsing script
Validating script
hello world
res63: org.apache.sysml.api.mlcontext.MLResults =
NoneMatrixMetadata
When supplying matrix data to Apache SystemML using the MLContext API, matrix metadata can be
supplied using a MatrixMetadata object. Supplying characteristics about a matrix can significantly
improve performance. For some types of input matrices, supplying metadata is mandatory.
Metadata at a minimum typically consists of the number of rows and columns in
a matrix. The number of non-zeros can also be supplied.
Additionally, the number of rows and columns per block can be supplied, although in typical usage it’s probably fine to use the default values used by SystemML (1,000 rows and 1,000 columns per block). SystemML handles a matrix internally by splitting the matrix into chunks, or blocks. The number of rows and columns per block refers to the size of these matrix blocks.
CSV RDD with No Metadata:
Here we see an example of inputting an RDD of Strings in CSV format with no metadata. Note that in general it is recommended that metadata is supplied. We output the sum and mean of the cells in the matrix.
val rddCSV = sc.parallelize(Array("1.0,2.0", "3.0,4.0"))
val sumAndMean = dml("sum = sum(m); mean = mean(m)").in("m", rddCSV).out("sum", "mean")
ml.execute(sumAndMean)scala> val rddCSV = sc.parallelize(Array("1.0,2.0", "3.0,4.0"))
rddCSV: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[190] at parallelize at <console>:38
scala> val sumAndMean = dml("sum = sum(m); mean = mean(m)").in("m", rddCSV).out("sum", "mean")
sumAndMean: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (RDD) m: ParallelCollectionRDD[190] at parallelize at <console>:38
Outputs:
[1] sum
[2] mean
scala> ml.execute(sumAndMean)
res20: org.apache.sysml.api.mlcontext.MLResults =
[1] (Double) sum: 10.0
[2] (Double) mean: 2.5IJV RDD with Metadata:
Next, we’ll supply an RDD in IJV format. IJV is a sparse format where each line has three space-separated values. The first value indicates the row number, the second value indicates the column number, and the third value indicates the cell value. Since the total numbers of rows and columns can’t be determined from these IJV rows, we need to supply metadata describing the matrix size.
Here, we specify that our matrix has 3 rows and 3 columns.
val rddIJV = sc.parallelize(Array("1 1 1", "2 1 2", "1 2 3", "3 3 4"))
val mm3x3 = new MatrixMetadata(MatrixFormat.IJV, 3, 3)
val sumAndMean = dml("sum = sum(m); mean = mean(m)").in("m", rddIJV, mm3x3).out("sum", "mean")
ml.execute(sumAndMean)scala> val rddIJV = sc.parallelize(Array("1 1 1", "2 1 2", "1 2 3", "3 3 4"))
rddIJV: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[202] at parallelize at <console>:38
scala> val mm3x3 = new MatrixMetadata(MatrixFormat.IJV, 3, 3)
mm3x3: org.apache.sysml.api.mlcontext.MatrixMetadata = rows: 3, columns: 3, non-zeros: None, rows per block: None, columns per block: None
scala> val sumAndMean = dml("sum = sum(m); mean = mean(m)").in("m", rddIJV, mm3x3).out("sum", "mean")
sumAndMean: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (RDD) m: ParallelCollectionRDD[202] at parallelize at <console>:38
Outputs:
[1] sum
[2] mean
scala> ml.execute(sumAndMean)
res21: org.apache.sysml.api.mlcontext.MLResults =
[1] (Double) sum: 10.0
[2] (Double) mean: 1.1111111111111112Next, we’ll run the same DML, but this time we’ll specify that the input matrix is 4x4 instead of 3x3.
val rddIJV = sc.parallelize(Array("1 1 1", "2 1 2", "1 2 3", "3 3 4"))
val mm4x4 = new MatrixMetadata(MatrixFormat.IJV, 4, 4)
val sumAndMean = dml("sum = sum(m); mean = mean(m)").in("m", rddIJV, mm4x4).out("sum", "mean")
ml.execute(sumAndMean)scala> val rddIJV = sc.parallelize(Array("1 1 1", "2 1 2", "1 2 3", "3 3 4"))
rddIJV: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[210] at parallelize at <console>:38
scala> val mm4x4 = new MatrixMetadata(MatrixFormat.IJV, 4, 4)
mm4x4: org.apache.sysml.api.mlcontext.MatrixMetadata = rows: 4, columns: 4, non-zeros: None, rows per block: None, columns per block: None
scala> val sumAndMean = dml("sum = sum(m); mean = mean(m)").in("m", rddIJV, mm4x4).out("sum", "mean")
sumAndMean: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (RDD) m: ParallelCollectionRDD[210] at parallelize at <console>:38
Outputs:
[1] sum
[2] mean
scala> ml.execute(sumAndMean)
res22: org.apache.sysml.api.mlcontext.MLResults =
[1] (Double) sum: 10.0
[2] (Double) mean: 0.625Matrix Data Conversions and Performance
Internally, Apache SystemML uses a binary-block matrix representation, where a matrix is represented as a grouping of blocks. Each block is equal in size to the other blocks in the matrix and consists of a number of rows and columns. The default block size is 1,000 rows by 1,000 columns.
Conversion of a large set of data to a SystemML matrix representation can potentially be time-consuming. Therefore, if you use a set of data multiple times, one way to potentially improve performance is to convert it to a SystemML matrix representation and then use this representation rather than performing the data conversion each time.
There are currently two mechanisms for this in SystemML: (1) BinaryBlockMatrix and (2) Matrix.
BinaryBlockMatrix:
If you have an input DataFrame, it can be converted to a BinaryBlockMatrix, and this BinaryBlockMatrix can be passed as an input rather than passing in the DataFrame as an input.
For example, suppose we had a 10000x1000 matrix represented as a DataFrame, as we saw in an earlier example. Now suppose we create two Script objects with the DataFrame as an input, as shown below. In the Spark Shell, when executing this code, you can see that each of the two Script object creations requires the time-consuming data conversion step.
import org.apache.spark.sql._
import org.apache.spark.sql.types.{StructType,StructField,DoubleType}
import scala.util.Random
val numRows = 10000
val numCols = 1000
val data = sc.parallelize(0 to numRows-1).map { _ => Row.fromSeq(Seq.fill(numCols)(Random.nextDouble)) }
val schema = StructType((0 to numCols-1).map { i => StructField("C" + i, DoubleType, true) } )
val df = sqlContext.createDataFrame(data, schema)
val mm = new MatrixMetadata(numRows, numCols)
val minMaxMeanScript = dml(minMaxMean).in("Xin", df, mm).out("minOut", "maxOut", "meanOut")
val minMaxMeanScript = dml(minMaxMean).in("Xin", df, mm).out("minOut", "maxOut", "meanOut")Rather than passing in a DataFrame each time to the Script object creation, let’s instead create a BinaryBlockMatrix object based on the DataFrame and pass this BinaryBlockMatrix to the Script object creation. If we run the code below in the Spark Shell, we see that the data conversion step occurs when the BinaryBlockMatrix object is created. However, when we create a Script object twice, we see that no conversion penalty occurs, since this conversion occurred when the BinaryBlockMatrix was created.
import org.apache.spark.sql._
import org.apache.spark.sql.types.{StructType,StructField,DoubleType}
import scala.util.Random
val numRows = 10000
val numCols = 1000
val data = sc.parallelize(0 to numRows-1).map { _ => Row.fromSeq(Seq.fill(numCols)(Random.nextDouble)) }
val schema = StructType((0 to numCols-1).map { i => StructField("C" + i, DoubleType, true) } )
val df = sqlContext.createDataFrame(data, schema)
val mm = new MatrixMetadata(numRows, numCols)
val bbm = new BinaryBlockMatrix(df, mm)
val minMaxMeanScript = dml(minMaxMean).in("Xin", bbm).out("minOut", "maxOut", "meanOut")
val minMaxMeanScript = dml(minMaxMean).in("Xin", bbm).out("minOut", "maxOut", "meanOut")Matrix:
When a matrix is returned as an output, it is returned as a Matrix object, which is a wrapper around a SystemML MatrixObject. As a result, an output Matrix is already in a SystemML representation, meaning that it can be passed as an input with no data conversion penalty.
As an example, here we read in matrix x as an RDD in CSV format. We create a Script that adds one to all
values in the matrix. We obtain the resulting matrix y as a Matrix. We execute the
script five times, feeding the output matrix as the input matrix for the next script execution.
val rddCSV = sc.parallelize(Array("1.0,2.0", "3.0,4.0"))
val add = dml("y = x + 1").in("x", rddCSV).out("y")
for (i <- 1 to 5) {
println("#" + i + ":");
val m = ml.execute(add).getMatrix("y")
m.toRDDStringCSV.collect.foreach(println)
add.in("x", m)
}scala> val rddCSV = sc.parallelize(Array("1.0,2.0", "3.0,4.0"))
rddCSV: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[341] at parallelize at <console>:53
scala> val add = dml("y = x + 1").in("x", rddCSV).out("y")
add: org.apache.sysml.api.mlcontext.Script =
Inputs:
[1] (RDD) x: ParallelCollectionRDD[341] at parallelize at <console>:53
Outputs:
[1] y
scala> for (i <- 1 to 5) {
| println("#" + i + ":");
| val m = ml.execute(add).getMatrix("y")
| m.toRDDStringCSV.collect.foreach(println)
| add.in("x", m)
| }
#1:
2.0,3.0
4.0,5.0
#2:
3.0,4.0
5.0,6.0
#3:
4.0,5.0
6.0,7.0
#4:
5.0,6.0
7.0,8.0
#5:
6.0,7.0
8.0,9.0Jupyter (PySpark) Notebook Example - Poisson Nonnegative Matrix Factorization
Similar to the Scala API, SystemML also provides a Python MLContext API. Before usage, you’ll need to install it first.
Here, we’ll explore the use of SystemML via PySpark in a Jupyter notebook. This Jupyter notebook example can be nicely viewed in a rendered state on GitHub, and can be downloaded here to a directory of your choice.
From the directory with the downloaded notebook, start Jupyter with PySpark:
PYSPARK_DRIVER_PYTHON=jupyter PYSPARK_DRIVER_PYTHON_OPTS="notebook" pyspark
PYSPARK_PYTHON=python3 PYSPARK_DRIVER_PYTHON=jupyter PYSPARK_DRIVER_PYTHON_OPTS="notebook" pyspark
This will open Jupyter in a browser:
We can then open up the SystemML-PySpark-Recommendation-Demo notebook.
Set up the notebook and download the data
%load_ext autoreload
%autoreload 2
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from systemml import MLContext, dml # pip install systeml
plt.rcParams['figure.figsize'] = (10, 6)%%sh
# Download dataset
curl -O http://snap.stanford.edu/data/amazon0601.txt.gz
gunzip amazon0601.txt.gzUse PySpark to load the data in as a Spark DataFrame
# Load data
import pyspark.sql.functions as F
dataPath = "amazon0601.txt"
X_train = (sc.textFile(dataPath)
.filter(lambda l: not l.startswith("#"))
.map(lambda l: l.split("\t"))
.map(lambda prods: (int(prods[0]), int(prods[1]), 1.0))
.toDF(("prod_i", "prod_j", "x_ij"))
.filter("prod_i < 500 AND prod_j < 500") # Filter for memory constraints
.cache())
max_prod_i = X_train.select(F.max("prod_i")).first()[0]
max_prod_j = X_train.select(F.max("prod_j")).first()[0]
numProducts = max(max_prod_i, max_prod_j) + 1 # 0-based indexing
print("Total number of products: {}".format(numProducts))Create a SystemML MLContext object
# Create SystemML MLContext
ml = MLContext(sc)Define a kernel for Poisson nonnegative matrix factorization (PNMF) in DML
# Define PNMF kernel in SystemML's DSL using the R-like syntax for PNMF
pnmf = """
# data & args
X = X+1 # change product IDs to be 1-based, rather than 0-based
V = table(X[,1], X[,2])
size = ifdef($size, -1)
if(size > -1) {
V = V[1:size,1:size]
}
n = nrow(V)
m = ncol(V)
range = 0.01
W = Rand(rows=n, cols=rank, min=0, max=range, pdf="uniform")
H = Rand(rows=rank, cols=m, min=0, max=range, pdf="uniform")
losses = matrix(0, rows=max_iter, cols=1)
# run PNMF
i=1
while(i <= max_iter) {
# update params
H = (H * (t(W) %*% (V/(W%*%H))))/t(colSums(W))
W = (W * ((V/(W%*%H)) %*% t(H)))/t(rowSums(H))
# compute loss
losses[i,] = -1 * (sum(V*log(W%*%H)) - as.scalar(colSums(W)%*%rowSums(H)))
i = i + 1;
}
"""Execute the algorithm
# Run the PNMF script on SystemML with Spark
script = dml(pnmf).input(X=X_train, max_iter=100, rank=10).output("W", "H", "losses")
W, H, losses = ml.execute(script).get("W", "H", "losses")Retrieve the losses during training and plot them
# Plot training loss over time
xy = losses.toDF().sort("__INDEX").map(lambda r: (r[0], r[1])).collect()
x, y = zip(*xy)
plt.plot(x, y)
plt.xlabel('Iteration')
plt.ylabel('Loss')
plt.title('PNMF Training Loss')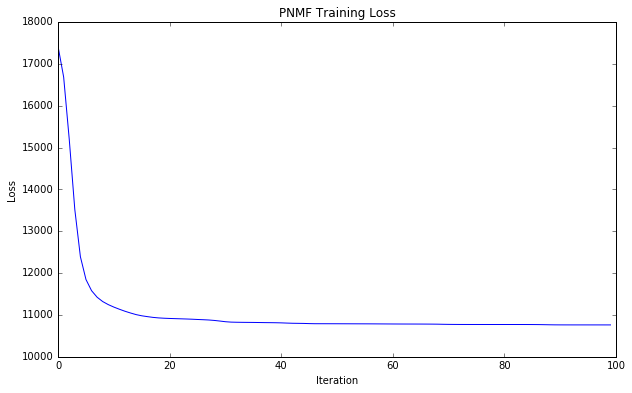
Spark Shell Example - OLD API
** NOTE: This API is old and has been deprecated. **
Please use the new MLContext API instead.
Start Spark Shell with SystemML
To use SystemML with the Spark Shell, the SystemML jar can be referenced using the Spark Shell’s --jars option.
Instructions to build the SystemML jar can be found in the SystemML GitHub README.
./bin/spark-shell --executor-memory 4G --driver-memory 4G --jars SystemML.jarHere is an example of Spark Shell with SystemML and YARN.
./bin/spark-shell --master yarn-client --num-executors 3 --driver-memory 5G --executor-memory 5G --executor-cores 4 --jars SystemML.jarCreate MLContext
An MLContext object can be created by passing its constructor a reference to the SparkContext.
scala>import org.apache.sysml.api.MLContext
import org.apache.sysml.api.MLContext
scala> val ml = new MLContext(sc)
ml: org.apache.sysml.api.MLContext = org.apache.sysml.api.MLContext@33e38c6bimport org.apache.sysml.api.MLContext
val ml = new MLContext(sc)Create DataFrame
For demonstration purposes, we’ll create a DataFrame consisting of 100,000 rows and 1,000 columns
of random doubles.
scala> import org.apache.spark.sql._
import org.apache.spark.sql._
scala> import org.apache.spark.sql.types.{StructType,StructField,DoubleType}
import org.apache.spark.sql.types.{StructType, StructField, DoubleType}
scala> import scala.util.Random
import scala.util.Random
scala> val numRows = 100000
numRows: Int = 100000
scala> val numCols = 1000
numCols: Int = 1000
scala> val data = sc.parallelize(0 to numRows-1).map { _ => Row.fromSeq(Seq.fill(numCols)(Random.nextDouble)) }
data: org.apache.spark.rdd.RDD[org.apache.spark.sql.Row] = MapPartitionsRDD[1] at map at <console>:33
scala> val schema = StructType((0 to numCols-1).map { i => StructField("C" + i, DoubleType, true) } )
schema: org.apache.spark.sql.types.StructType = StructType(StructField(C0,DoubleType,true), StructField(C1,DoubleType,true), StructField(C2,DoubleType,true), StructField(C3,DoubleType,true), StructField(C4,DoubleType,true), StructField(C5,DoubleType,true), StructField(C6,DoubleType,true), StructField(C7,DoubleType,true), StructField(C8,DoubleType,true), StructField(C9,DoubleType,true), StructField(C10,DoubleType,true), StructField(C11,DoubleType,true), StructField(C12,DoubleType,true), StructField(C13,DoubleType,true), StructField(C14,DoubleType,true), StructField(C15,DoubleType,true), StructField(C16,DoubleType,true), StructField(C17,DoubleType,true), StructField(C18,DoubleType,true), StructField(C19,DoubleType,true), StructField(C20,DoubleType,true), StructField(C21,DoubleType,true), ...
scala> val df = sqlContext.createDataFrame(data, schema)
df: org.apache.spark.sql.DataFrame = [C0: double, C1: double, C2: double, C3: double, C4: double, C5: double, C6: double, C7: double, C8: double, C9: double, C10: double, C11: double, C12: double, C13: double, C14: double, C15: double, C16: double, C17: double, C18: double, C19: double, C20: double, C21: double, C22: double, C23: double, C24: double, C25: double, C26: double, C27: double, C28: double, C29: double, C30: double, C31: double, C32: double, C33: double, C34: double, C35: double, C36: double, C37: double, C38: double, C39: double, C40: double, C41: double, C42: double, C43: double, C44: double, C45: double, C46: double, C47: double, C48: double, C49: double, C50: double, C51: double, C52: double, C53: double, C54: double, C55: double, C56: double, C57: double, C58: double, C5...import org.apache.spark.sql._
import org.apache.spark.sql.types.{StructType,StructField,DoubleType}
import scala.util.Random
val numRows = 100000
val numCols = 1000
val data = sc.parallelize(0 to numRows-1).map { _ => Row.fromSeq(Seq.fill(numCols)(Random.nextDouble)) }
val schema = StructType((0 to numCols-1).map { i => StructField("C" + i, DoubleType, true) } )
val df = sqlContext.createDataFrame(data, schema)Helper Methods
For convenience, we’ll create some helper methods. The SystemML output data is encapsulated in
an MLOutput object. The getScalar() method extracts a scalar value from a DataFrame returned by
MLOutput. The getScalarDouble() method returns such a value as a Double, and the
getScalarInt() method returns such a value as an Int.
scala> import org.apache.sysml.api.MLOutput
import org.apache.sysml.api.MLOutput
scala> def getScalar(outputs: MLOutput, symbol: String): Any =
| outputs.getDF(sqlContext, symbol).first()(1)
getScalar: (outputs: org.apache.sysml.api.MLOutput, symbol: String)Any
scala> def getScalarDouble(outputs: MLOutput, symbol: String): Double =
| getScalar(outputs, symbol).asInstanceOf[Double]
getScalarDouble: (outputs: org.apache.sysml.api.MLOutput, symbol: String)Double
scala> def getScalarInt(outputs: MLOutput, symbol: String): Int =
| getScalarDouble(outputs, symbol).toInt
getScalarInt: (outputs: org.apache.sysml.api.MLOutput, symbol: String)Intimport org.apache.sysml.api.MLOutput
def getScalar(outputs: MLOutput, symbol: String): Any =
outputs.getDF(sqlContext, symbol).first()(1)
def getScalarDouble(outputs: MLOutput, symbol: String): Double =
getScalar(outputs, symbol).asInstanceOf[Double]
def getScalarInt(outputs: MLOutput, symbol: String): Int =
getScalarDouble(outputs, symbol).toIntConvert DataFrame to Binary-Block Matrix
SystemML is optimized to operate on a binary-block format for matrix representation. For large datasets, conversion from DataFrame to binary-block can require a significant quantity of time. Explicit DataFrame to binary-block conversion allows algorithm performance to be measured separately from data conversion time.
The SystemML binary-block matrix representation can be thought of as a two-dimensional array of blocks, where each block
consists of a number of rows and columns. In this example, we specify a matrix consisting
of blocks of size 1000x1000. The experimental dataFrameToBinaryBlock() method of RDDConverterUtilsExt is used
to convert the DataFrame df to a SystemML binary-block matrix, which is represented by the datatype
JavaPairRDD[MatrixIndexes, MatrixBlock].
scala> import org.apache.sysml.runtime.instructions.spark.utils.{RDDConverterUtilsExt => RDDConverterUtils}
import org.apache.sysml.runtime.instructions.spark.utils.{RDDConverterUtilsExt=>RDDConverterUtils}
scala> import org.apache.sysml.runtime.matrix.MatrixCharacteristics;
import org.apache.sysml.runtime.matrix.MatrixCharacteristics
scala> val numRowsPerBlock = 1000
numRowsPerBlock: Int = 1000
scala> val numColsPerBlock = 1000
numColsPerBlock: Int = 1000
scala> val mc = new MatrixCharacteristics(numRows, numCols, numRowsPerBlock, numColsPerBlock)
mc: org.apache.sysml.runtime.matrix.MatrixCharacteristics = [100000 x 1000, nnz=-1, blocks (1000 x 1000)]
scala> val sysMlMatrix = RDDConverterUtils.dataFrameToBinaryBlock(sc, df, mc, false)
sysMlMatrix: org.apache.spark.api.java.JavaPairRDD[org.apache.sysml.runtime.matrix.data.MatrixIndexes,org.apache.sysml.runtime.matrix.data.MatrixBlock] = org.apache.spark.api.java.JavaPairRDD@2bce3248import org.apache.sysml.runtime.instructions.spark.utils.{RDDConverterUtilsExt => RDDConverterUtils}
import org.apache.sysml.runtime.matrix.MatrixCharacteristics;
val numRowsPerBlock = 1000
val numColsPerBlock = 1000
val mc = new MatrixCharacteristics(numRows, numCols, numRowsPerBlock, numColsPerBlock)
val sysMlMatrix = RDDConverterUtils.dataFrameToBinaryBlock(sc, df, mc, false)DML Script
For this example, we will utilize the following DML Script called shape.dml that reads in a matrix and outputs the number of rows and the
number of columns, each represented as a matrix.
X = read($Xin)
m = matrix(nrow(X), rows=1, cols=1)
n = matrix(ncol(X), rows=1, cols=1)
write(m, $Mout)
write(n, $Nout)Execute Script
Let’s execute our DML script, as shown in the example below. The call to reset() of MLContext is not necessary here, but this method should
be called if you need to reset inputs and outputs or if you would like to call execute() with a different script.
An example of registering the DataFrame df as an input to the X variable is shown but commented out. If a DataFrame is registered directly,
it will implicitly be converted to SystemML’s binary-block format. However, since we’ve already explicitly converted the DataFrame to the
binary-block fixed variable systemMlMatrix, we will register this input to the X variable. We register the m and n variables
as outputs.
When SystemML is executed via DMLScript (such as in Standalone Mode), inputs are supplied as either command-line named arguments
or positional argument. These inputs are specified in DML scripts by prepending them with a $. Values are read from or written
to files using read/write (DML) and load/save (PyDML) statements. When utilizing the MLContext API,
inputs and outputs can be other data representations, such as DataFrames. The input and output data are bound to DML variables.
The named arguments in the shape.dml script do not have default values set for them, so we create a Map to map the required named
arguments to blank Strings so that the script can pass validation.
The shape.dml script is executed by the call to execute(), where we supply the Map of required named arguments. The
execution results are returned as the MLOutput fixed variable outputs. The number of rows is obtained by calling the getStaticInt()
helper method with the outputs object and "m". The number of columns is retrieved by calling getStaticInt() with
outputs and "n".
scala> ml.reset()
scala> //ml.registerInput("X", df) // implicit conversion of DataFrame to binary-block
scala> ml.registerInput("X", sysMlMatrix, numRows, numCols)
scala> ml.registerOutput("m")
scala> ml.registerOutput("n")
scala> val nargs = Map("Xin" -> " ", "Mout" -> " ", "Nout" -> " ")
nargs: scala.collection.immutable.Map[String,String] = Map(Xin -> " ", Mout -> " ", Nout -> " ")
scala> val outputs = ml.execute("shape.dml", nargs)
15/10/12 16:29:15 WARN : Your hostname, derons-mbp.usca.ibm.com resolves to a loopback/non-reachable address: 127.0.0.1, but we couldn't find any external IP address!
15/10/12 16:29:15 WARN OptimizerUtils: Auto-disable multi-threaded text read for 'text' and 'csv' due to thread contention on JRE < 1.8 (java.version=1.7.0_80).
outputs: org.apache.sysml.api.MLOutput = org.apache.sysml.api.MLOutput@4d424743
scala> val m = getScalarInt(outputs, "m")
m: Int = 100000
scala> val n = getScalarInt(outputs, "n")
n: Int = 1000ml.reset()
//ml.registerInput("X", df) // implicit conversion of DataFrame to binary-block
ml.registerInput("X", sysMlMatrix, numRows, numCols)
ml.registerOutput("m")
ml.registerOutput("n")
val nargs = Map("Xin" -> " ", "Mout" -> " ", "Nout" -> " ")
val outputs = ml.execute("shape.dml", nargs)
val m = getScalarInt(outputs, "m")
val n = getScalarInt(outputs, "n")DML Script as String
The MLContext API allows a DML script to be specified
as a String. Here, we specify a DML script as a fixed String variable called minMaxMeanScript.
This DML will find the minimum, maximum, and mean value of a matrix.
scala> val minMaxMeanScript: String =
| """
| Xin = read(" ")
| minOut = matrix(min(Xin), rows=1, cols=1)
| maxOut = matrix(max(Xin), rows=1, cols=1)
| meanOut = matrix(mean(Xin), rows=1, cols=1)
| write(minOut, " ")
| write(maxOut, " ")
| write(meanOut, " ")
| """
minMaxMeanScript: String =
"
Xin = read(" ")
minOut = matrix(min(Xin), rows=1, cols=1)
maxOut = matrix(max(Xin), rows=1, cols=1)
meanOut = matrix(mean(Xin), rows=1, cols=1)
write(minOut, " ")
write(maxOut, " ")
write(meanOut, " ")
"val minMaxMeanScript: String =
"""
Xin = read(" ")
minOut = matrix(min(Xin), rows=1, cols=1)
maxOut = matrix(max(Xin), rows=1, cols=1)
meanOut = matrix(mean(Xin), rows=1, cols=1)
write(minOut, " ")
write(maxOut, " ")
write(meanOut, " ")
"""Scala Wrapper for DML
We can create a Scala wrapper for our invocation of the minMaxMeanScript DML String. The minMaxMean() method
takes a JavaPairRDD[MatrixIndexes, MatrixBlock] parameter, which is a SystemML binary-block matrix representation.
It also takes a rows parameter indicating the number of rows in the matrix, a cols parameter indicating the number
of columns in the matrix, and an MLContext parameter. The minMaxMean() method
returns a tuple consisting of the minimum value in the matrix, the maximum value in the matrix, and the computed
mean value of the matrix.
scala> import org.apache.sysml.runtime.matrix.data.MatrixIndexes
import org.apache.sysml.runtime.matrix.data.MatrixIndexes
scala> import org.apache.sysml.runtime.matrix.data.MatrixBlock
import org.apache.sysml.runtime.matrix.data.MatrixBlock
scala> import org.apache.spark.api.java.JavaPairRDD
import org.apache.spark.api.java.JavaPairRDD
scala> def minMaxMean(mat: JavaPairRDD[MatrixIndexes, MatrixBlock], rows: Int, cols: Int, ml: MLContext): (Double, Double, Double) = {
| ml.reset()
| ml.registerInput("Xin", mat, rows, cols)
| ml.registerOutput("minOut")
| ml.registerOutput("maxOut")
| ml.registerOutput("meanOut")
| val outputs = ml.executeScript(minMaxMeanScript)
| val minOut = getScalarDouble(outputs, "minOut")
| val maxOut = getScalarDouble(outputs, "maxOut")
| val meanOut = getScalarDouble(outputs, "meanOut")
| (minOut, maxOut, meanOut)
| }
minMaxMean: (mat: org.apache.spark.api.java.JavaPairRDD[org.apache.sysml.runtime.matrix.data.MatrixIndexes,org.apache.sysml.runtime.matrix.data.MatrixBlock], rows: Int, cols: Int, ml: org.apache.sysml.api.MLContext)(Double, Double, Double)import org.apache.sysml.runtime.matrix.data.MatrixIndexes
import org.apache.sysml.runtime.matrix.data.MatrixBlock
import org.apache.spark.api.java.JavaPairRDD
def minMaxMean(mat: JavaPairRDD[MatrixIndexes, MatrixBlock], rows: Int, cols: Int, ml: MLContext): (Double, Double, Double) = {
ml.reset()
ml.registerInput("Xin", mat, rows, cols)
ml.registerOutput("minOut")
ml.registerOutput("maxOut")
ml.registerOutput("meanOut")
val outputs = ml.executeScript(minMaxMeanScript)
val minOut = getScalarDouble(outputs, "minOut")
val maxOut = getScalarDouble(outputs, "maxOut")
val meanOut = getScalarDouble(outputs, "meanOut")
(minOut, maxOut, meanOut)
}Invoking DML via Scala Wrapper
Here, we invoke minMaxMeanScript using our minMaxMean() Scala wrapper method. It returns a tuple
consisting of the minimum value in the matrix, the maximum value in the matrix, and the mean value of the matrix.
scala> val (min, max, mean) = minMaxMean(sysMlMatrix, numRows, numCols, ml)
15/10/13 14:33:11 WARN OptimizerUtils: Auto-disable multi-threaded text read for 'text' and 'csv' due to thread contention on JRE < 1.8 (java.version=1.7.0_80).
min: Double = 5.378949397005783E-9
max: Double = 0.9999999934660398
mean: Double = 0.499988222338507val (min, max, mean) = minMaxMean(sysMlMatrix, numRows, numCols, ml)Zeppelin Notebook Example - Linear Regression Algorithm - OLD API
** NOTE: This API is old and has been deprecated. **
Please use the new MLContext API instead.
Next, we’ll consider an example of a SystemML linear regression algorithm run from Spark through an Apache Zeppelin notebook. Instructions to clone and build Zeppelin can be found at the GitHub Apache Zeppelin site. This example also will look at the Spark ML linear regression algorithm.
This Zeppelin notebook example can be imported by choosing Import note -> Add from URL from the Zeppelin main page, then insert the following URL:
https://raw.githubusercontent.com/apache/incubator-systemml/master/samples/zeppelin-notebooks/2AZ2AQ12B/note.json
Alternatively download note.json, then import it by choosing Import note -> Choose a JSON here from the Zeppelin main page.
A conf/zeppelin-env.sh file is created based on conf/zeppelin-env.sh.template. For
this demonstration, it features SPARK_HOME, SPARK_SUBMIT_OPTIONS, and ZEPPELIN_SPARK_USEHIVECONTEXT
environment variables:
export SPARK_HOME=/Users/example/spark-1.5.1-bin-hadoop2.6
export SPARK_SUBMIT_OPTIONS="--jars /Users/example/systemml/system-ml/target/SystemML.jar"
export ZEPPELIN_SPARK_USEHIVECONTEXT=false
Start Zeppelin using the zeppelin.sh script:
bin/zeppelin.sh
After opening Zeppelin in a brower, we see the “SystemML - Linear Regression” note in the list of available Zeppelin notes.
If we go to the “SystemML - Linear Regression” note, we see that the note consists of several cells of code.
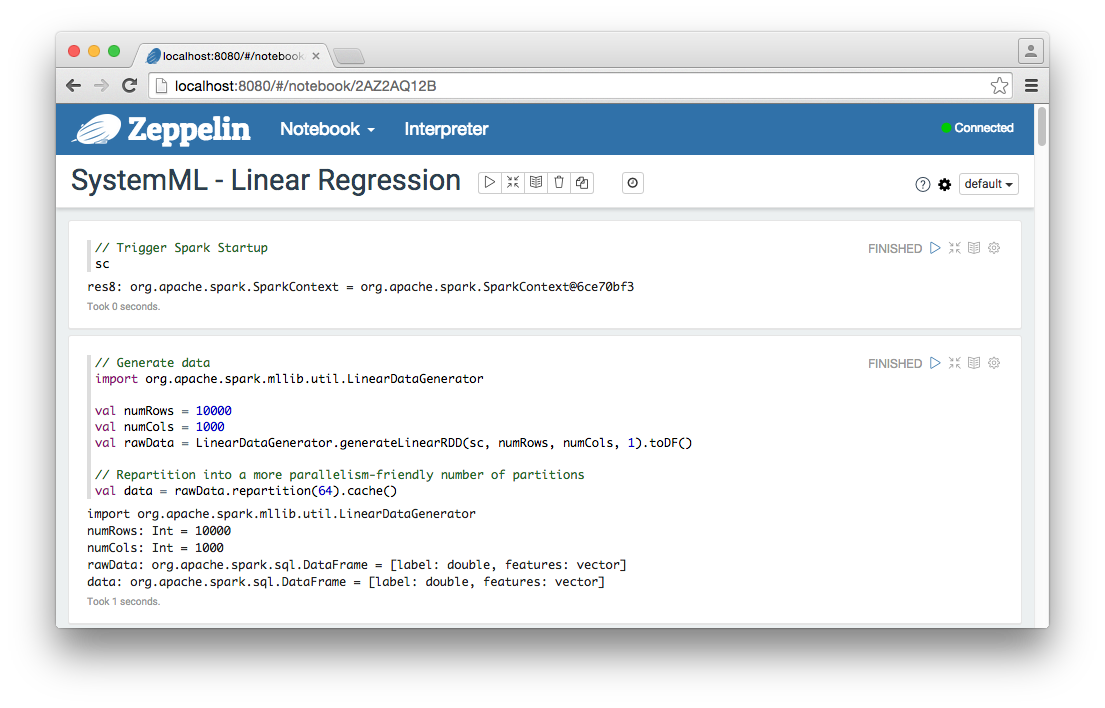
Let’s briefly consider these cells.
Trigger Spark Startup
This cell triggers Spark to initialize by calling the SparkContext sc object. Information regarding these startup operations can be viewed in the
console window in which zeppelin.sh is running.
Cell:
// Trigger Spark Startup
scOutput:
res8: org.apache.spark.SparkContext = org.apache.spark.SparkContext@6ce70bf3Generate Linear Regression Test Data
The Spark LinearDataGenerator is used to generate test data for the Spark ML and SystemML linear regression algorithms.
Cell:
// Generate data
import org.apache.spark.mllib.util.LinearDataGenerator
import sqlContext.implicits._
val numRows = 10000
val numCols = 1000
val rawData = LinearDataGenerator.generateLinearRDD(sc, numRows, numCols, 1).toDF()
// Repartition into a more parallelism-friendly number of partitions
val data = rawData.repartition(64).cache()Output:
import org.apache.spark.mllib.util.LinearDataGenerator
numRows: Int = 10000
numCols: Int = 1000
rawData: org.apache.spark.sql.DataFrame = [label: double, features: vector]
data: org.apache.spark.sql.DataFrame = [label: double, features: vector]Train using Spark ML Linear Regression Algorithm for Comparison
For purpose of comparison, we can train a model using the Spark ML linear regression algorithm.
Cell:
// Spark ML
import org.apache.spark.ml.regression.LinearRegression
// Model Settings
val maxIters = 100
val reg = 0
val elasticNetParam = 0 // L2 reg
// Fit the model
val lr = new LinearRegression()
.setMaxIter(maxIters)
.setRegParam(reg)
.setElasticNetParam(elasticNetParam)
val start = System.currentTimeMillis()
val model = lr.fit(data)
val trainingTime = (System.currentTimeMillis() - start).toDouble / 1000.0
// Summarize the model over the training set and gather some metrics
val trainingSummary = model.summary
val r2 = trainingSummary.r2
val iters = trainingSummary.totalIterations
val trainingTimePerIter = trainingTime / itersOutput:
import org.apache.spark.ml.regression.LinearRegression
maxIters: Int = 100
reg: Int = 0
elasticNetParam: Int = 0
lr: org.apache.spark.ml.regression.LinearRegression = linReg_a7f51d676562
start: Long = 1444672044647
model: org.apache.spark.ml.regression.LinearRegressionModel = linReg_a7f51d676562
trainingTime: Double = 12.985
trainingSummary: org.apache.spark.ml.regression.LinearRegressionTrainingSummary = org.apache.spark.ml.regression.LinearRegressionTrainingSummary@227ba28b
r2: Double = 0.9677118209276552
iters: Int = 17
trainingTimePerIter: Double = 0.7638235294117647Spark ML Linear Regression Summary Statistics
Summary statistics for the Spark ML linear regression algorithm are displayed by this cell.
Cell:
// Print statistics
println(s"R2: ${r2}")
println(s"Iterations: ${iters}")
println(s"Training time per iter: ${trainingTimePerIter} seconds")Output:
R2: 0.9677118209276552
Iterations: 17
Training time per iter: 0.7638235294117647 secondsSystemML Linear Regression Algorithm
The linearReg fixed String variable is set to
a linear regression algorithm written in DML, SystemML’s Declarative Machine Learning language.
Cell:
// SystemML kernels
val linearReg =
"""
#
# THIS SCRIPT SOLVES LINEAR REGRESSION USING THE CONJUGATE GRADIENT ALGORITHM
#
# INPUT PARAMETERS:
# --------------------------------------------------------------------------------------------
# NAME TYPE DEFAULT MEANING
# --------------------------------------------------------------------------------------------
# X String --- Matrix X of feature vectors
# Y String --- 1-column Matrix Y of response values
# icpt Int 0 Intercept presence, shifting and rescaling the columns of X:
# 0 = no intercept, no shifting, no rescaling;
# 1 = add intercept, but neither shift nor rescale X;
# 2 = add intercept, shift & rescale X columns to mean = 0, variance = 1
# reg Double 0.000001 Regularization constant (lambda) for L2-regularization; set to nonzero
# for highly dependend/sparse/numerous features
# tol Double 0.000001 Tolerance (epsilon); conjugate graduent procedure terminates early if
# L2 norm of the beta-residual is less than tolerance * its initial norm
# maxi Int 0 Maximum number of conjugate gradient iterations, 0 = no maximum
# --------------------------------------------------------------------------------------------
#
# OUTPUT:
# B Estimated regression parameters (the betas) to store
#
# Note: Matrix of regression parameters (the betas) and its size depend on icpt input value:
# OUTPUT SIZE: OUTPUT CONTENTS: HOW TO PREDICT Y FROM X AND B:
# icpt=0: ncol(X) x 1 Betas for X only Y ~ X %*% B[1:ncol(X), 1], or just X %*% B
# icpt=1: ncol(X)+1 x 1 Betas for X and intercept Y ~ X %*% B[1:ncol(X), 1] + B[ncol(X)+1, 1]
# icpt=2: ncol(X)+1 x 2 Col.1: betas for X & intercept Y ~ X %*% B[1:ncol(X), 1] + B[ncol(X)+1, 1]
# Col.2: betas for shifted/rescaled X and intercept
#
fileX = "";
fileY = "";
fileB = "";
intercept_status = ifdef ($icpt, 0); # $icpt=0;
tolerance = ifdef ($tol, 0.000001); # $tol=0.000001;
max_iteration = ifdef ($maxi, 0); # $maxi=0;
regularization = ifdef ($reg, 0.000001); # $reg=0.000001;
X = read (fileX);
y = read (fileY);
n = nrow (X);
m = ncol (X);
ones_n = matrix (1, rows = n, cols = 1);
zero_cell = matrix (0, rows = 1, cols = 1);
# Introduce the intercept, shift and rescale the columns of X if needed
m_ext = m;
if (intercept_status == 1 | intercept_status == 2) # add the intercept column
{
X = append (X, ones_n);
m_ext = ncol (X);
}
scale_lambda = matrix (1, rows = m_ext, cols = 1);
if (intercept_status == 1 | intercept_status == 2)
{
scale_lambda [m_ext, 1] = 0;
}
if (intercept_status == 2) # scale-&-shift X columns to mean 0, variance 1
{ # Important assumption: X [, m_ext] = ones_n
avg_X_cols = t(colSums(X)) / n;
var_X_cols = (t(colSums (X ^ 2)) - n * (avg_X_cols ^ 2)) / (n - 1);
is_unsafe = ppred (var_X_cols, 0.0, "<=");
scale_X = 1.0 / sqrt (var_X_cols * (1 - is_unsafe) + is_unsafe);
scale_X [m_ext, 1] = 1;
shift_X = - avg_X_cols * scale_X;
shift_X [m_ext, 1] = 0;
} else {
scale_X = matrix (1, rows = m_ext, cols = 1);
shift_X = matrix (0, rows = m_ext, cols = 1);
}
# Henceforth, if intercept_status == 2, we use "X %*% (SHIFT/SCALE TRANSFORM)"
# instead of "X". However, in order to preserve the sparsity of X,
# we apply the transform associatively to some other part of the expression
# in which it occurs. To avoid materializing a large matrix, we rewrite it:
#
# ssX_A = (SHIFT/SCALE TRANSFORM) %*% A --- is rewritten as:
# ssX_A = diag (scale_X) %*% A;
# ssX_A [m_ext, ] = ssX_A [m_ext, ] + t(shift_X) %*% A;
#
# tssX_A = t(SHIFT/SCALE TRANSFORM) %*% A --- is rewritten as:
# tssX_A = diag (scale_X) %*% A + shift_X %*% A [m_ext, ];
lambda = scale_lambda * regularization;
beta_unscaled = matrix (0, rows = m_ext, cols = 1);
if (max_iteration == 0) {
max_iteration = m_ext;
}
i = 0;
# BEGIN THE CONJUGATE GRADIENT ALGORITHM
r = - t(X) %*% y;
if (intercept_status == 2) {
r = scale_X * r + shift_X %*% r [m_ext, ];
}
p = - r;
norm_r2 = sum (r ^ 2);
norm_r2_initial = norm_r2;
norm_r2_target = norm_r2_initial * tolerance ^ 2;
while (i < max_iteration & norm_r2 > norm_r2_target)
{
if (intercept_status == 2) {
ssX_p = scale_X * p;
ssX_p [m_ext, ] = ssX_p [m_ext, ] + t(shift_X) %*% p;
} else {
ssX_p = p;
}
q = t(X) %*% (X %*% ssX_p);
if (intercept_status == 2) {
q = scale_X * q + shift_X %*% q [m_ext, ];
}
q = q + lambda * p;
a = norm_r2 / sum (p * q);
beta_unscaled = beta_unscaled + a * p;
r = r + a * q;
old_norm_r2 = norm_r2;
norm_r2 = sum (r ^ 2);
p = -r + (norm_r2 / old_norm_r2) * p;
i = i + 1;
}
# END THE CONJUGATE GRADIENT ALGORITHM
if (intercept_status == 2) {
beta = scale_X * beta_unscaled;
beta [m_ext, ] = beta [m_ext, ] + t(shift_X) %*% beta_unscaled;
} else {
beta = beta_unscaled;
}
# Output statistics
avg_tot = sum (y) / n;
ss_tot = sum (y ^ 2);
ss_avg_tot = ss_tot - n * avg_tot ^ 2;
var_tot = ss_avg_tot / (n - 1);
y_residual = y - X %*% beta;
avg_res = sum (y_residual) / n;
ss_res = sum (y_residual ^ 2);
ss_avg_res = ss_res - n * avg_res ^ 2;
R2_temp = 1 - ss_res / ss_avg_tot
R2 = matrix(R2_temp, rows=1, cols=1)
write(R2, "")
totalIters = matrix(i, rows=1, cols=1)
write(totalIters, "")
# Prepare the output matrix
if (intercept_status == 2) {
beta_out = append (beta, beta_unscaled);
} else {
beta_out = beta;
}
write (beta_out, fileB);
"""Output:
None
Helper Methods
This cell contains helper methods to return Double and Int values from output generated by the MLContext API.
Cell:
// Helper functions
import org.apache.sysml.api.MLOutput
def getScalar(outputs: MLOutput, symbol: String): Any =
outputs.getDF(sqlContext, symbol).first()(1)
def getScalarDouble(outputs: MLOutput, symbol: String): Double =
getScalar(outputs, symbol).asInstanceOf[Double]
def getScalarInt(outputs: MLOutput, symbol: String): Int =
getScalarDouble(outputs, symbol).toIntOutput:
import org.apache.sysml.api.MLOutput
getScalar: (outputs: org.apache.sysml.api.MLOutput, symbol: String)Any
getScalarDouble: (outputs: org.apache.sysml.api.MLOutput, symbol: String)Double
getScalarInt: (outputs: org.apache.sysml.api.MLOutput, symbol: String)IntConvert DataFrame to Binary-Block Format
SystemML uses a binary-block format for matrix data representation. This cell
explicitly converts the DataFrame data object to a binary-block features matrix
and single-column label matrix, both represented by the
JavaPairRDD[MatrixIndexes, MatrixBlock] datatype.
Cell:
// Imports
import org.apache.sysml.api.MLContext
import org.apache.sysml.runtime.instructions.spark.utils.{RDDConverterUtilsExt => RDDConverterUtils}
import org.apache.sysml.runtime.matrix.MatrixCharacteristics;
// Create SystemML context
val ml = new MLContext(sc)
// Convert data to proper format
val mcX = new MatrixCharacteristics(numRows, numCols, 1000, 1000)
val mcY = new MatrixCharacteristics(numRows, 1, 1000, 1000)
val X = RDDConverterUtils.vectorDataFrameToBinaryBlock(sc, data, mcX, false, "features")
val y = RDDConverterUtils.dataFrameToBinaryBlock(sc, data.select("label"), mcY, false)
// val y = data.select("label")
// Cache
val X2 = X.cache()
val y2 = y.cache()
val cnt1 = X2.count()
val cnt2 = y2.count()Output:
import org.apache.sysml.api.MLContext
import org.apache.sysml.runtime.instructions.spark.utils.{RDDConverterUtilsExt=>RDDConverterUtils}
import org.apache.sysml.runtime.matrix.MatrixCharacteristics
ml: org.apache.sysml.api.MLContext = org.apache.sysml.api.MLContext@38d59245
mcX: org.apache.sysml.runtime.matrix.MatrixCharacteristics = [10000 x 1000, nnz=-1, blocks (1000 x 1000)]
mcY: org.apache.sysml.runtime.matrix.MatrixCharacteristics = [10000 x 1, nnz=-1, blocks (1000 x 1000)]
X: org.apache.spark.api.java.JavaPairRDD[org.apache.sysml.runtime.matrix.data.MatrixIndexes,org.apache.sysml.runtime.matrix.data.MatrixBlock] = org.apache.spark.api.java.JavaPairRDD@b5a86e3
y: org.apache.spark.api.java.JavaPairRDD[org.apache.sysml.runtime.matrix.data.MatrixIndexes,org.apache.sysml.runtime.matrix.data.MatrixBlock] = org.apache.spark.api.java.JavaPairRDD@56377665
X2: org.apache.spark.api.java.JavaPairRDD[org.apache.sysml.runtime.matrix.data.MatrixIndexes,org.apache.sysml.runtime.matrix.data.MatrixBlock] = org.apache.spark.api.java.JavaPairRDD@650f29d2
y2: org.apache.spark.api.java.JavaPairRDD[org.apache.sysml.runtime.matrix.data.MatrixIndexes,org.apache.sysml.runtime.matrix.data.MatrixBlock] = org.apache.spark.api.java.JavaPairRDD@334857a8
cnt1: Long = 10
cnt2: Long = 10Train using SystemML Linear Regression Algorithm
Now, we can train our model using the SystemML linear regression algorithm. We register the features matrix X and the label matrix y as inputs. We register the beta_out matrix,
R2, and totalIters as outputs.
Cell:
// Register inputs & outputs
ml.reset()
ml.registerInput("X", X, numRows, numCols)
ml.registerInput("y", y, numRows, 1)
// ml.registerInput("y", y)
ml.registerOutput("beta_out")
ml.registerOutput("R2")
ml.registerOutput("totalIters")
// Run the script
val start = System.currentTimeMillis()
val outputs = ml.executeScript(linearReg)
val trainingTime = (System.currentTimeMillis() - start).toDouble / 1000.0
// Get outputs
val B = outputs.getDF(sqlContext, "beta_out").sort("ID").drop("ID")
val r2 = getScalarDouble(outputs, "R2")
val iters = getScalarInt(outputs, "totalIters")
val trainingTimePerIter = trainingTime / itersOutput:
start: Long = 1444672090620
outputs: org.apache.sysml.api.MLOutput = org.apache.sysml.api.MLOutput@5d2c22d0
trainingTime: Double = 1.176
B: org.apache.spark.sql.DataFrame = [C1: double]
r2: Double = 0.9677079547216473
iters: Int = 12
trainingTimePerIter: Double = 0.09799999999999999SystemML Linear Regression Summary Statistics
SystemML linear regression summary statistics are displayed by this cell.
Cell:
// Print statistics
println(s"R2: ${r2}")
println(s"Iterations: ${iters}")
println(s"Training time per iter: ${trainingTimePerIter} seconds")
B.describe().show()Output:
R2: 0.9677079547216473
Iterations: 12
Training time per iter: 0.2334166666666667 seconds
+-------+-------------------+
|summary| C1|
+-------+-------------------+
| count| 1000|
| mean| 0.0184500840658385|
| stddev| 0.2764750319432085|
| min|-0.5426068958986378|
| max| 0.5225309861616542|
+-------+-------------------+Jupyter (PySpark) Notebook Example - Poisson Nonnegative Matrix Factorization - OLD API
** NOTE: This API is old and has been deprecated. **
Please use the new MLContext API instead.
Here, we’ll explore the use of SystemML via PySpark in a Jupyter notebook. This Jupyter notebook example can be nicely viewed in a rendered state on GitHub, and can be downloaded here to a directory of your choice.
From the directory with the downloaded notebook, start Jupyter with PySpark:
PYSPARK_DRIVER_PYTHON=jupyter PYSPARK_DRIVER_PYTHON_OPTS="notebook" $SPARK_HOME/bin/pyspark --master local[*] --driver-class-path $SYSTEMML_HOME/SystemML.jar
This will open Jupyter in a browser:

We can then open up the SystemML-PySpark-Recommendation-Demo notebook:
Set up the notebook and download the data
%load_ext autoreload
%autoreload 2
%matplotlib inline
# Add SystemML PySpark API file.
sc.addPyFile("https://raw.githubusercontent.com/apache/incubator-systemml/3d5f9b11741f6d6ecc6af7cbaa1069cde32be838/src/main/java/org/apache/sysml/api/python/SystemML.py")
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (10, 6)%%sh
# Download dataset
curl -O http://snap.stanford.edu/data/amazon0601.txt.gz
gunzip amazon0601.txt.gzUse PySpark to load the data in as a Spark DataFrame
# Load data
import pyspark.sql.functions as F
dataPath = "amazon0601.txt"
X_train = (sc.textFile(dataPath)
.filter(lambda l: not l.startswith("#"))
.map(lambda l: l.split("\t"))
.map(lambda prods: (int(prods[0]), int(prods[1]), 1.0))
.toDF(("prod_i", "prod_j", "x_ij"))
.filter("prod_i < 500 AND prod_j < 500") # Filter for memory constraints
.cache())
max_prod_i = X_train.select(F.max("prod_i")).first()[0]
max_prod_j = X_train.select(F.max("prod_j")).first()[0]
numProducts = max(max_prod_i, max_prod_j) + 1 # 0-based indexing
print("Total number of products: {}".format(numProducts))Create a SystemML MLContext object
# Create SystemML MLContext
from SystemML import MLContext
ml = MLContext(sc)Define a kernel for Poisson nonnegative matrix factorization (PNMF) in DML
# Define PNMF kernel in SystemML's DSL using the R-like syntax for PNMF
pnmf = """
# data & args
X = read($X)
X = X+1 # change product IDs to be 1-based, rather than 0-based
V = table(X[,1], X[,2])
size = ifdef($size, -1)
if(size > -1) {
V = V[1:size,1:size]
}
max_iteration = as.integer($maxiter)
rank = as.integer($rank)
n = nrow(V)
m = ncol(V)
range = 0.01
W = Rand(rows=n, cols=rank, min=0, max=range, pdf="uniform")
H = Rand(rows=rank, cols=m, min=0, max=range, pdf="uniform")
losses = matrix(0, rows=max_iteration, cols=1)
# run PNMF
i=1
while(i <= max_iteration) {
# update params
H = (H * (t(W) %*% (V/(W%*%H))))/t(colSums(W))
W = (W * ((V/(W%*%H)) %*% t(H)))/t(rowSums(H))
# compute loss
losses[i,] = -1 * (sum(V*log(W%*%H)) - as.scalar(colSums(W)%*%rowSums(H)))
i = i + 1;
}
# write outputs
write(losses, $lossout)
write(W, $Wout)
write(H, $Hout)
"""Execute the algorithm
# Run the PNMF script on SystemML with Spark
ml.reset()
outputs = ml.executeScript(pnmf, {"X": X_train, "maxiter": 100, "rank": 10}, ["W", "H", "losses"])Retrieve the losses during training and plot them
# Plot training loss over time
losses = outputs.getDF(sqlContext, "losses")
xy = losses.sort(losses.ID).map(lambda r: (r[0], r[1])).collect()
x, y = zip(*xy)
plt.plot(x, y)
plt.xlabel('Iteration')
plt.ylabel('Loss')
plt.title('PNMF Training Loss')
Recommended Spark Configuration Settings
For best performance, we recommend setting the following flags when running SystemML with Spark:
--conf spark.driver.maxResultSize=0 --conf spark.akka.frameSize=128.